INTRODUCTIONOtitis media with effusion (OME) is an inflammation of the middle ear in which there is a collection of retro-tympanic liquid without signs or symptoms of acute infection and with intact tympanic membrane. The terms secretory otitis media, non-suppurative otitis media, serous otitis media and mucoid otitis media are used as synonym of otitis media with effusion, but they are not as precise. The characterization of type of effusion can be hindered by opacification and edema of the tympanic membrane 1.
OME is normally considered a direct continuum of the inflammatory process that occurs during prolonged or recurrent episodes of acute otitis media (AOM), which is confirmed not only by the fact that almost all cases of OME follow an episode of AOM, but also based on animal experiments 2, 3.
The infectious origin of OME is supported by this fact. Conversely, only 20-40% of the cases of OME have positive culture. Streptococcus pneumoniae, Haemophilus influenzae and Moraxella catarrhalis are the most frequently detected microorganism 4-7. Recently, some Polymerase Chain Reaction techniques (PCR) were adapted to detect bacterial DNA in middle ear effusion and its use has increased to 80% the frequency of positive results to microorganisms in studied samples 8-11.
Alloiococcus otitidis was recently referred as the bacterium involved in the genesis of OME. This germ presents slow growth and it is hardly ever detected by conventional culture technique, but it was identified in 20-50% of the middle ear aspirates of patients with OME investigated with PCR 10-12.
The need to previously define which germs are going to be looked for by PCR and the prevalence above referred have defined the option to investigate S. pneumoniae, H. influenzae, M. catarrhalis and A. otitidis.
The identification of prevalence of microorganisms responsible or involved in the cases of OME can help in selecting the most appropriate antimicrobial agent, minimizing the complications that require surgeries. Therefore, we tried to determine the prevalence of bacteria previously referred to in middle ear effusion of children with OME (recurrent otitis media and chronic otitis media with effusion) using culture analysis and PCR technique.
MATERIAL AND METHODWe conducted a transversal, observation, modern study of sub-individual data (ears). Based on the expected prevalence of positive bacterial results of 65% by PCR and 25% by culture analysis, with 10% error margin, it was estimated that the minimum sample would require 72 effusion samples. Since we wanted to compare the distribution of positive responses in cases of recurrent otitis media (ROM) and chronic otitis media with effusion (OMEC) we decided to have a sample with 120 effusion collections. It allowed comparison of the difference of at least 20% between the groups, fixing it at ? = 0.05 and ? = 0.20.
Seventy-five children with diagnosis of OME coming from the ENT pediatric unit of Porto Alegre, were enrolled in the study during the period between June 2001 and October 2002. It included patients that were aged between 11 months and 10 years, with otitis media effusion for six weeks or more (OME), diagnosis of recurrent OM (three or more episodes of AOM within 6 months), or OMEC (persistent effusion for over 3 months) that presented indication to myringotomy and ventilation tube placement. The main investigator clinically followed up the patients at least for 6 weeks before the surgery. Immittanciometry was conducted, whenever necessary, to confirm presence of effusion in recurrent otitis media and in all patients with diagnosis of chronic otitis media with effusion, who underwent audiometry and immittanciometry. They also conducted pneumatic otoscopy under videoendoscopic view in all patients 24 hours before the surgical procedure to confirm absence of upper airway infections. Patients that presented AOM, other upper airway infections, use of antibiotics or history of completion of treatment only 7 days before surgery were excluded from the study.
The surgery was conducted under general anesthesia. The material used was aspirated from the middle ear with a collector by Alden Senturia (Alden-Senturia collector®, Storz Instruments, St. Louis, USA), after cleaning and removing the cerumen from the external auditory canal (EAC), antiseptic cleaning of EAC with alcohol at 70% and tympanocentesis on the anterior-inferior quadrant of the tympanic membrane. There was no indication for use of means of transportation, since the material was sent to culture and PCR analysis within maximum 15 minutes after collection.
The PCR technique used in this study is a method for simultaneous detection (multiplex PCR) of S. pneumoniae, H. influenzae, M. catarrhalis and A. otitidis10. Gene 16S rRNA, which contains both variable and constant sequences, was chosen as the amplification goal of the PCR. The constant sequences are common to many bacteria and the variable sequences are specific to each species. The effusion samples previously frozen were later defrozen and DNA was extracted using the commercially available kit QIAamp® (Qiagen, Valencia, USA). For each reaction, we added 1.25U polymerase Taq (GeneAmp®, Applyed Biosystems, Branchburg, USA) with the appropriate buffer, as instructed in the kit. The amplified products were separated into agarose gel at 3% and visualized under ultraviolet light. Positive or negative results for each bacteria were then reported 10, 11.
For culture purposes, the material was processed in plaques that contained sheep blood agar and chocolate agar (Biolab-Mérieux, Rio de Janeiro, Brazil) and then incubated in aerobiose during 24 hours at 37oC. The bacterial identification was conducted by automation (Vitek®, bioMérieux S.A., Marcy-l'Etoile, France). If there was no bacterial growth during this time, the plaques were reincubated for another 96 hours before being released with the definitely negative result.
The results of the culture and PCR analysis were described by absolute and percentage frequency. The impact of the contribution of the PCR over the results of the culture was assessed by a percentage delta (?%), defined as:
?% = final value - initial value x 100
initial value
The associations of matched observations were assessed using chi-square test (?2) by McNemar and the independent observations used ?2 test with Yates correction and Fisher's exact test. The adopted level of significance was = 0.05.
The protocol was approved by the Research Ethics Committee, Research and Post-Graduation Group, Hospital de Clinicas, Porto Alegre. All patients signed the informed consent term as a condition for their enrollment in the study.
RESULTSWe analyzed the effusion samples of 128 middle ears. All the 75 children included in the study provided material for the analysis. The ages ranged from 11 months to 9 years and 4 months (mean ± standard deviation = 34.7±18.5 months), 60% were male subjects and they were all Caucasians.
Fifty-three of them (70.7%) of the 75 patients had secretion from both ears and in 29 cases (54.7%), we found different bacteria in each ear, when analyzing it by PCR. We diagnosed ROM in 69.3% and OMEC in 30.7% of the patients. The patients with ROM had mean of 5.3±1.4 episodes of otitis every 6 months and those with OMEC had mean duration of effusion of 4.8±1.1 months.
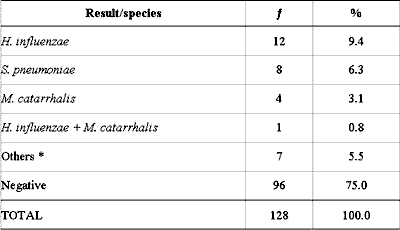
The culture identified bacteria in 32 (25.1%) of the 128 middle ear effusion samples and the main pathogens (S. pneumoniae, H. influenzae and M. catarrhalis) were isolated in 25 (19.6%) of them. A. otitidis was not detected through culture, and bacteria normally considered of low clinical importance or non-pathogenic in the middle ear were found in 7 cases (5.5%) (Table 1).
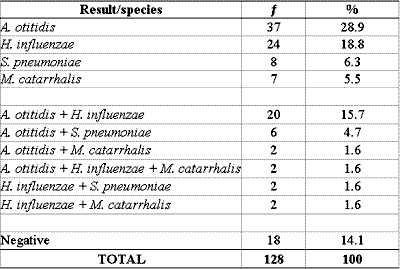
PCR was positive for one or more bacteria in 110 (85.9%) of the 128 effusion samples collected. In 76 samples, there was identification of only one agent and in 34 samples we found a mix of bacterial DNA. A. otitidis was found in 67 (52.3%), H. influenzae in 50 (39.1%), S. pneumoniae in 16 (12.5%) and M. catarrhalis in 13 (10.2%) of the 128 samples (Tables 2 and 3).
There was better performance of PCR when compared to culture in detecting bacteria in the studied samples. The difference between the proportion of positive effusions in PCR and in culture was statistically significant for all studied bacteria, individually and collectively (Table 3). The increase observed in the proportion of effusion with positive test in the culture caused by PCR (?%) is presented in Table 4. The increment in data obtained by PCR represents an increase of 340% of the total number of effusion samples identified as positive for one studied germ, or an increase of 192% when excluding the positive PCR tests for A. otitidis.
Out of 128 effusion samples, 87 (68.0%) were obtained from patients with ROM, and 41 (31%) from children with OMEC. By PCR, 78 (89.7%) of the 87 effusion samples were from ROM patients and 32 (78%) of the 41 samples from OMEC patients were positive for one or more of the four studied bacteria. There was no statistically significant difference between the positive levels (Table 5).
We also conducted an analysis of the results obtained with PCR in effusion samples of children from the two different age ranges, younger and older than 2 years, and it showed similar frequency in all bacteria in both groups, except for H. influenzae, found in 52.4% of the effusion samples of children below 2 years, and in 32.6% of the older children (P=0.049) (Table 6).
DISCUSSIONIn the past, OME was seen as a strictly inflammatory process and effusion was considered sterile. However, in 1958, Senturia et al. 13 found bacteria in samples of OME effusion and changed the concepts valid at the time.
Lack of uniformity in defining OME and duration of effusion and lack of criteria related to use of antibiotics by patients that have effusion are the hurdles for correct valuation of information present in the literature and for comparison of data generated by the study. We understand that the difficulty to interpret middle ear effusion bacteriological data results from the fact, in some occasions, that it is almost impossible to determine in which stage of the continuum of otitis media the patients are. It is known that studies that investigate characteristics of prolonged effusion in middle ear use aspirated material during myringotomy and later when ventilation tubes are placed. The criteria to conduct these surgical procedures with aspiration of effusion seem to be relatively uniform, but there are few publications that clearly inform about duration of effusion and concomitant use of antibiotics.
Regular monitoring of the child by the investigator in order to characterize the type of OM and to follow up the duration of effusion is a difficult task to healthcare outpatients units. Therefore, our study included only patients from the pediatric unit of the main investigator, being the only responsible person for initial assessment and follow up, which included pneumatic otoscopy under endoscopic view in each visit.
The culture analysis provided similar results to those reported in the international literature concerning type and frequency of bacteria present in effusion of patients with OME (20-40%) 4-7. The higher prevalence of H. influenzae, followed by S. pneumoniae and M. catarrhalis is quite similar to that from other studies, which also showed an inversion in order between pneumococcus and hemophilus, when compared to those obtained from patients with AOM 6. Individual percentages for H. influenzae (10.2%), S. pneumoniae (6.3%) and M. catarrhalis (3.9%) are quite similar to those recently detected by Sutton et al.7 upon analyzing the samples of ROM and OMEC which were 12.1%, 9.6% and 5.6%, respectively.
The comparison of the present data with those reported by some Brazilian studies presents discrepancies. The results ranged from negative responses in all culture tests to 33.6% positive response with the method 14-16. Among the predominant microorganism, we can include, in addition to H. influenzae, S. pneumoniae and M. catarrhalis (not necessarily in this order), S. aureus, S. epidermidis and P. aeruginosa, which were among the most prevalent ones 15,16. It is likely that the differences in inclusion criteria, microbiology methodology, geographical variation and variability influenced by reduced size of the sample are responsible for the disparities observed when comparing the data of these studies and the correlation with the present study.
We did not identify A. otitidis through culture, confirming its characteristics of slow and difficult growth and, in accordance with the studies published in Europe and Japan, in which it was also impossible to identify this germ by culture 10-12, 17, 18. It is important to refer to BHI (brain heart infusion), a culture mean known as the most appropriate to detect A. otitidis, which was not used in the present study because of technical difficulties, which resulted in a limitation. Alternatively, the culture means explored were the same as those that provided "accidental growth" that allowed Faden and Dryja 19 in 1989 to first identify this microorganism.
As to PCR, it increased significantly the frequency of bacterial identification (85.9%) and it is in accordance with the literature, in which the percentage ranged from 50 to 94.5% 8-12, 18. The frequency with which each bacterium was identified pointed towards higher prevalence of A. otitidis (52.3%), followed by H. influenzae (39.1%), S. pneumoniae (12.5%) and M. catarrhalis (10.2%). Upon comparing these data to those presented by the already mentioned authors, it is detected that they fell within the intervals described for H. influenzae (12.5 - 70.2%) and S. pneumoniae (6.4 - 35.0%). Conversely, the percentages obtained in the present study were somewhat higher than those referred for A. otitidis (20 - 50%) and lower than those reported by M. catarrhalis (16 - 63%), without, however, characterizing a significant difference. We understand that the small differences detected can be easily explained by the variability of the sample. Therefore, there are no indicators in the findings of the present study that are effectively different from literature data. Lastly, since we did not find any study in Brazil that analyzed effusion of OME using PCR, we concluded that further studies should be performed, especially because Brazil is a very large country and counts on different population groups.
The comparison of results showed that all samples with positive results in culture also have positive response for the same bacteria in the PCR, and the difference between the proportion of positive effusion in culture (19.6%) and in PCR (85.9%) was statistically significant for the bacteria included in the study, both individually and collectively (P<0.01). These results are similar and equally significant to those in the international literature 8-10, 12, 18. Data collection obtained in PCR represents, compared to regular culture, an increase of 340% in the total number of effusion samples identified as positive for the studied microorganisms, results similar to the increase of 268% reported by Post et al. 9 and the 260% increase reported by Hendolin et al10.
The possibility of having positive PCR to represent residual DNA fragments, not necessarily indicative of the presence of viable pathogenic bacteria, is argued by some authors 20. Animal studies have clarified, in our opinion, the main questions of applicability of PCR in investigation of middle ear infectious diseases. Using the OM model in chinchilla, Post et al. 21, demonstrated that purified bacterial DNA and bacteria DNA inactivated by heat did not persist in the amplifiable form for more than 3 days after inoculation. Moreover, bacteria sensitive to antibiotics became non-growing after the third day of treatment, but DNA remained amplifiable for 3 weeks. Aul et al.22, supported by an animal model, showed that strains of hemophilus resistant to ampicillin remained identifiable both by PCR and culture for over one month, whereas DNA of inoculated pneumococcus in small number of colonies was amplified for 21 days, despite the fact that they were not culturally isolated after the 3rd day. As in the previous study, purified DNA of hemophilus and DNA of moraxella inactivated by heat did not persist beyond 2 days. Additional evidence was presented by Rayner et al.23, who identified mRNA of H. influenzae, a molecule with average life of seconds to minutes, in human effusion culturally negative for the same microorganism. Moreover, they demonstrated that the DNA of the same microorganism was not identified in the samples in which mRNA was amplified, suggesting that the identification technique of the former, more common, simple and economical was enough for diagnostic purposes. Therefore, we understand that it suggests that effusion has efficient mechanisms to remove non-viable bacteria, in addition to quick degrading DNA process. Conversely, these findings raise some hypotheses concerning the complex relationship that can define the middle ear, between the microbe and the host, unable to be detected using traditional culture techniques. Metabolically active bacteria would be capable of persisting in the middle ear and the lower bacterial identification rates by culture, when compared to PCR, would be explained by one or more of the hypotheses: (a) the amount of microorganism would be lower than the limits of detection of the culture, normally pointed as being 104 CFU/ml24; (b) germs would assume L forms, variant of bacteria that lose the capacity to synthesize the peptidoglycan layer on their walls, and therefore, change their original shape, becoming resistant to ?-lactam antibiotics and imposing restrictions to growing 25; (c) bacteria would assume biofilm form, a bacterial community in which they are bond to the surface or one to the others, resulting in lower metabolic activity and greater capability to resist antimicrobial agents 23. Based on electron microscopy, Ehrlich et al.26 has recently demonstrated the formation of biofilm on the mucosa of the middle ear in an animal model of otitis media and they suggested that the mechanism could be an important factor in the pathogenesis of the middle ear with chronic effusion. The three situations referred above could be caused by treatment with antibiotics and by immune response of the host. Moreover, microorganisms in L form or grouped as biofilm have lower metabolic and reproductive activity when compared to plan tonic bacteria, which would be at least partially responsible for chronic inflammation and persistence of middle ear effusion in children, without induction of exuberant symptomatology 23, 25, 26. Evidently, the confirmation of this hypothesis depends on the continuation of studies.
A. otitidis has deserved attention from those that investigate the possible bacterial causes of otitis media. Its frequent intracellular location and the associated presence of many inflammatory cells suggest the pathogenic role of the microorganism 10, 27, 28. Its identification with effusion of greater duration and mucoid appearance would be associated with more chronic stages of OME 18. From the pioneer publication by Faden and Dryja19 and the classification of the germ by Aguirre and Collins29, an expressive number of studies have demonstrated the significant prevalence of A. otitidis in patients, considering its evidence to classification as middle ear pathogen 10, 12, 18.
The use of molecular biology techniques, such as PCR, excluded the limitations of cultures and allowed better characterization of bacteria that are recently identified or not isolated by conventional methods. One justification for low rate of bacterial isolation in samples of OME effusion is the existence of slow and difficult to growth organisms, and it is the case of A. otitidis, possibly explaining why it was not described longer before 27,28.
Out of 128 studied effusion samples, 68% were from ROM patients and 32% from OMEC patients. One of the few studies that clearly defined inclusion criteria for both forms of OME was conducted by Sutton et al.7, who pointed out the percentages of 57.2% (ROM) and 42.8% (OMEC). In PCR analysis, A. otitidis and H. influenzae presented similar distribution in both groups. The former confirmed its higher prevalence in the whole study and the latter agreed with the literature, which states that among the three main middle ear pathogens, it is the most prevalence microorganism in OME. Conversely, S. pneumoniae was 7.2 times more frequent in ROM than in OMEC, which reached statistically significant difference.
The literature is not uniform in presenting data that can be compared to our study; Hendolin et al.11 did not refer different distribution of A. otitidis, Jero et al.30 and Leskinen et al.18 found it more frequently in effusion that had lasted for over 3 months (OMEC). Brook et al.31 found more pneumococcus (but also hemophilus) in those who had effusion for less time. It is possible to infer that those with ROM presented effusion for shorter time and had recent history of acute episodes, which can show bacteriological profile similar to that found in AOM, in which pneumococcus predominated. Brook et al. 31 stated that "it is confirmed that effusion in children with OME of shorter duration, when they have positive culture, present pathogens that are normally found in AOM, which may have implications to treatment if we believe that bacteria play a role in the pathology". It is understood that - excluding the information about prevalence of A. otitidis, whose importance in the pathogenesis of the middle ear disease still has to be confirmed - the data obtained suggest that we should consider the duration of effusion in the middle ear and the interval concerning the last episodes of AOM to make decisions about antibiotic therapy, if necessary.
Despite the fact that it is not the object of the present study, we compared the data obtained for children in two different age ranges. The group of children older than 2 years contributed with twice more samples than younger children. There was no different in frequency of bacteria in the two groups, except for H. influenzae, which was more frequently found in children below 2 years (P=0.049). Brook et al.31 and Jero and Karma32 employed conventional bacteriological methods and found statistically significant higher incidence of H. influenzae and S. pneumoniae in effusion of children younger than 2 years with OME, which, in the present study, was confirmed only in relation to hemophilus. Leskinen et al.18 used the PCR technique and described the prevalence of S. pneumoniae in the group of children younger than 2 years (P=0.04), without confirming this finding for other studied bacteria. Conversely, Riding et al.4 did not identify any age-related difference. If we accept the higher incidence of complications in otitis media in children younger than 2 years as a fact and understand that the peak of incidence is at the end of the first year of life, we can have a justification to efficiently and completely identify and characterize the disease in this age range, pointing to its similarities and differences when comparing to older children and adults, including bacteriological aspects.
We can conclude that the present study is the first conducted in Brazil in which prevalence of A. otitidis was specifically studied. The bacterium may be related to etiopathogenesis of OME, since it was the most frequently detected microorganism in this study and it was identified in 88% of the 34 samples in which the studied germs were associated, reinforcing the concept of supporting role that this bacterium can have on the pathogenic effect of other bacteria, possibly owing to the capacity of stimulating middle ear colonization 10, 18. Moreover, it justifies the conduction of studies about colonization sites and their characteristics of antimicrobial resistance.
CONCLUSIONSThe prevalence of OME bacteria in a group of Brazilian children is similar to that reported in other countries, being H. influenzae the most frequently found pathogen in middle ear. S. pneumoniae was more frequently detected in middle ear effusion of ROM than OMEC, showing statistically significant difference, which did not happen with the other studied bacteria. The highest frequency of pneumococcus in ROM suggested that bacteriology of OME of shorter duration is similar to that of AOM.
The results showed that PCR is more sensitive than culture to detect bacteria in the middle ear effusion and it is essential do detect A. otitidis.
The high percentage of A. otitidis detection suggests that further investigation should be performed about its role in the onset and prolongation of middle ear diseases.
ACKNOWLEDGEMENTWe would like to thank Prof Dr Manuel May Pereira for the microbiological support and to Laboratório Weinmann for having performed the culture and PCR analyses.
Table 1 - Culture analysis of 128 middle ear effusion collections.
: frequency
* Staphylococcus epidermidis (1), Streptococcus oralis (2), Brevibacterium sp (2) and
Corynebacterium auris (2).
Table 2 - PCR in samples of 128 middle ear effusion collections.
: frequency
PCR: Polymerase chain reaction
Table 3 - Comparison of culture and PCR in 128 effusion collections.
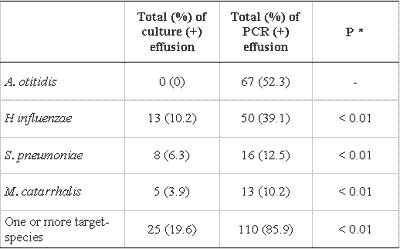
Data are presented as absolute and percentage figures.
* McNemar test
PCR: Polymerase chain reaction
Table 4 - Comparison of culture and PCR tests by studied germ.
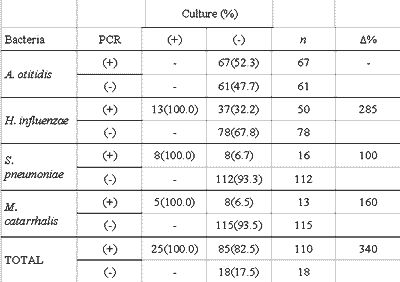
Data are presented as absolute values, percentages are calculated concerning the culture analysis.
n: number of effusions.
£%: percentage increase in positive results of culture, provided by the information from PCR
PCR: Polymerase chain reaction
Table 5 - Frequency of bacteria in 128 effusion samples of ROM and OMEC by PCR analysis.
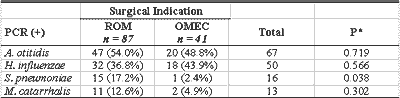
Data are presented as absolute and percentage values.
ROM: Recurrent otitis media; OMEC: otitis media with chronic effusion
PCR: Polymerase chain reaction
n: number of effusions
* X2 with Yates correction
Table 6 - Comparison of PCR results in 128 effusion samples of children younger and older than 2 years.
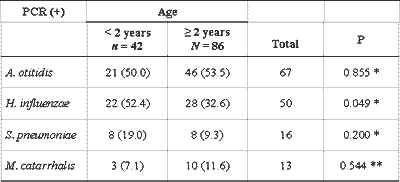
Data are presented as absolute and percentage numbers.
PCR: Polymerase chain reaction
* ?2 with Yates correction
** Fisher's exact test
n: number of effusions
Note: the adding up of positive effusions for each germ is greater than the total number of effusion collections (n), and the sum of the percentages is greater than 100 because of bacterial associations in the same collection.
1. Bluestone CD, Gates GA, Klein JO, Lim DJ, Mogi G, Ogra PL, et al. Recent advances in otitis media. 1. Definitions, terminology, and classification of otitis media. Ann Otol Rhinol Laryngol Suppl 2002; 188: 8-18. [Recent advances in otitis media: Report of the Seventh Research Conference].
2. Juhn S, Paparella M, Kim C, Goycoolea M, Giebink G. Pathogenesis of otitis media. Ann Otol Rhinol Laryngol 1977; 86: 481-92.
3. Giebink GS. Progress in understanding the pathophysiology of otitis media. Pediatr Rev 1989; 11: 133-8.
4. Riding KH, Bluestone CD, Michaels RH, Cantekin EI, Doyle WJ, Poziviak CS. Microbiology of recurrent and chronic otitis media with effusion. J Pediatr 1978; 93: 739-43.
5. Giebink GS, Juhn SK, Weber ML, Le CT. The bacteriology and cytology of chronic otitis media with effusion. Pediatr Infect Dis J 1982; 1: 98-103.
6. Bluestone CD, Stephenson JS, Martin LM. Ten-year review of otitis media pathogens. Pediatr Infect Dis J 1992; 11: S7-S11.
7. Sutton DV, Darrow DH, Derkay CS, Strasnick B. Resistant bacteria in middle ear fluid at the time of tympanostomy tube surgery. Ann Otol Rhinol Laryngol 2000; 109: 24-9.
8. Hotomi M, Tabata T, Kakiuchi H, Kunimoto M. Detection of Haemophilus influenzae in middle ear of otitis media with effusion by polymerase chain reaction. Int J Ped Otol 1993; 27: 119-126.
9. Post JC, Preston R, Aul J, Larkins-Pettigrew M, Rydquist-White J, Anderson K, et al. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA 1995; 273: 1598-1604.
10. Hendolin P, Markkanen A, Ylikoski J, Wahlfors J. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusion. J Clin Microbiol 1997; 35: 2854-8.
11. Hendolin P, Paulin L, Ylikoski J. Clinically applicable multiplex PCR for four middle ear pathogens. J Clin Microbiol 2000; 38: 125-32.
12. Beswick AJ, Lawley B, Fraise AP, Pahor AL, Brown NL. Detection of Alloiococcus otitis in mixed bacterial populations from middle-ear effusions of patients with otitis media. Lancet 1999; 354: 386-89.
13. Senturia BH, Gessert CF, Carr CD, Baumann ES. Studies concerned with tubotympanitis. Ann Otol Rhinol Laryngol 1958; 67: 440-67.
14. Saffer M, Lubianca Neto JF, Piltcher OB, Petrillo VF. Chronic secretory otitis media: negative bacteriology. Acta Otolaryngol 1996; 116: 836-9.
15. Filizzola VC, Weckx LL, Carlini D, Martino MD, Mimica IM. Estudo bacteriológico da secreção de orelha média em crianças com otite média secretora crônica. Rev Bras de Otorrinolaringol 1998; 64: 604-8.
16. Rezende VA, Almeida ER, Bento RF, Durigon EL, Botosso VF, Queiroz D. Estudo da flora bacteriana e viral na otite média secretora e rinofaringe na infância. Rev Bras Otorrinolaringol 1999; 65: 10-7.
17. Harimaya A, Fujii N, Mitsuzawa H, Yamazaki N, Himi T. Evaluation of specific antibody against Alloiococcus otitidis in the middle ear fluid. In: Takasaka T, Yuasa R, Hozawa K, editors. Recent advances in otitis media: Proceedings of Otitis Media 2001, April 16-20, Sendai, Japan. Bologna: Monduzzi/International Proceedings Division; 2001. p.151-4.
18. Leskinen K, Hendolin P, Virolainen-Julkunen A, Ylikoski J, Jero J. The clinical role of Alloiococcus otitidis in otitis media with effusion. Int J Pediatr Otorhinolaryngol 2002; 66: 41-8.
19. Faden H, Dryja D. Recovery of a unique bacterial organism in human middle ear fluid and its possible role in chronic otitis media. J Clin Microbiol 1989; 27: 2488-91.
20. Cantekin EI. Bacterial DNA fragments in otitis media with effusion. JAMA 1996; 275: 186.
21. Post JC, Aul JJ, White GJ, Wadowsky RM, Zavoral T, Tabari R, et al. PCR-based detection of bacterial DNA after antimicrobial treatment is indicative of persistent, viable bacteria in the chinchilla model of otitis media. Am J Otolaryngol 1996; 17: 106-11.
22. Aul JJ, Anderson KW, Wadowsky RM, Doyle WJ, Kingsley LA, Post JC, et al. Comparative evaluation of culture and PCR for the detection and determination of persistence of bacterial strains and DNAs in the Chinchilla laniger model of otitis media. Ann Otol Rhinol Laryngol 1998; 107: 508-13.
23. Rayner M, Zhang Y, Gorry M, Chen Y, Post J, Ehrlich G. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 1998; 279: 296-9.
24. Stenfors L, Räisänen S. How long do middle ear pathogens survive in mucoid effusion material? Acta Otolaryngol 1989; 107: 244-8.
25. Göksu N, Ataoglu H, Kemaloglu YK, Ataoglu Ö, Özsökmen D, Akyildiz N. Experimental otitis media induced by coagulase negative staphylococcus and its L-forms. Int J Ped Otorhinolaryngol 1996; 37: 201-16.
26. Ehrlich GD, Veeh R, Wang X, Costerton JW, Hayes JD, Hu FZ, et al. Mucosal biofilm formation on middle-ear effusion in the chinchilla model of otitis media. JAMA 2002; 287: 1710-5.
27. Bosley GS, Whitney A, Pruckler J, Moss W, Daneshvar M, Sih T, et al. Characterization of ear fluid isolates of Alloiococcus otitidis from patients with recurrent otitis media. J Clin Microbiol 1995; 33: 2876-80.
28. Sih T, Schwartz G, Bosley G, Facklam R. Chronic otitis media with effusion caused by Alloiococcus otitidis: clinical and laboratory features. In: American Society of Microbiology. Thirty Second Interscience Conference on Antimicrobial Agents and Chemotherapy. Washington: The Society; 1992. p. 222.
29. Aguirre M, Collins MD. Phylogenetic analysis of Alloiococcus otitis gen. nov., sp. nov., an organism from human middle ear fluid. Int J Syst Bacteriol 1992; 42: 79-83.
30. Jero J, Leskinen K, Virolainen A, Hendolin P, Ylikoski J. The role of Alloiococcus otitidis in the continuum of otitis media in children. Recent advances in otitis media: Seventh International Symposium: Fort Lauderdale, Fla, 1999. p. 195. [Abstracts]
31. Brook I, Yocum P, Shah K, Feldman B, Epstein S. Microbiology of serous otitis media in children: correlation with age and length of effusion. Ann Otol Rhinol Laryngol 2001; 110: 87-90.
32. Jero J, Karma P. Bacteriological findings and persistence of middle ear effusion in otitis media with effusion. Acta Otolaryngol Suppl 1997; 529: 22-6.
1 Otorhinolaryngologist, Master in Pediatrics, Federal University of Rio Grande do Sul, Fellow in Pediatric Otorhinolaryngology, University of Manitoba, Winnipeg, Canada.
2 Ph.D. in Molecular and Cellular Biology, University of Osaka, Japan. Responsible for the Division of Molecular Biology, Laboratório Weinmann, Porto Alegre.
3 Undergraduate, Medical School, Pontifícia Universidade Católica do Rio Grande do Sul.
4 Assistant Professor, Ph.D., Department of Otorhinolaryngology and Ophthalmology, Medical School, Federal University of Rio Grande do Sul.
Affiliation: Medical School, Federal University of Rio Grande do Sul, Porto Alegre, RS.
Address correspondence to: M. Beatriz Rotta Pereira. Rua Padre Chagas 415, cj. 902. Porto Alegre. 90570-080.
Tel (55 51) 3222-8909 - Fax (55 51) 3395-4666 - E-mail: bearotta@hotmail.com.
Study presented at III Congresso Triológico de Otorrinolaringologia (classified among those awarded Best Free Communication in Otology), October 8 - 11, 2003, Rio de Janeiro, RJ, Brazil.


